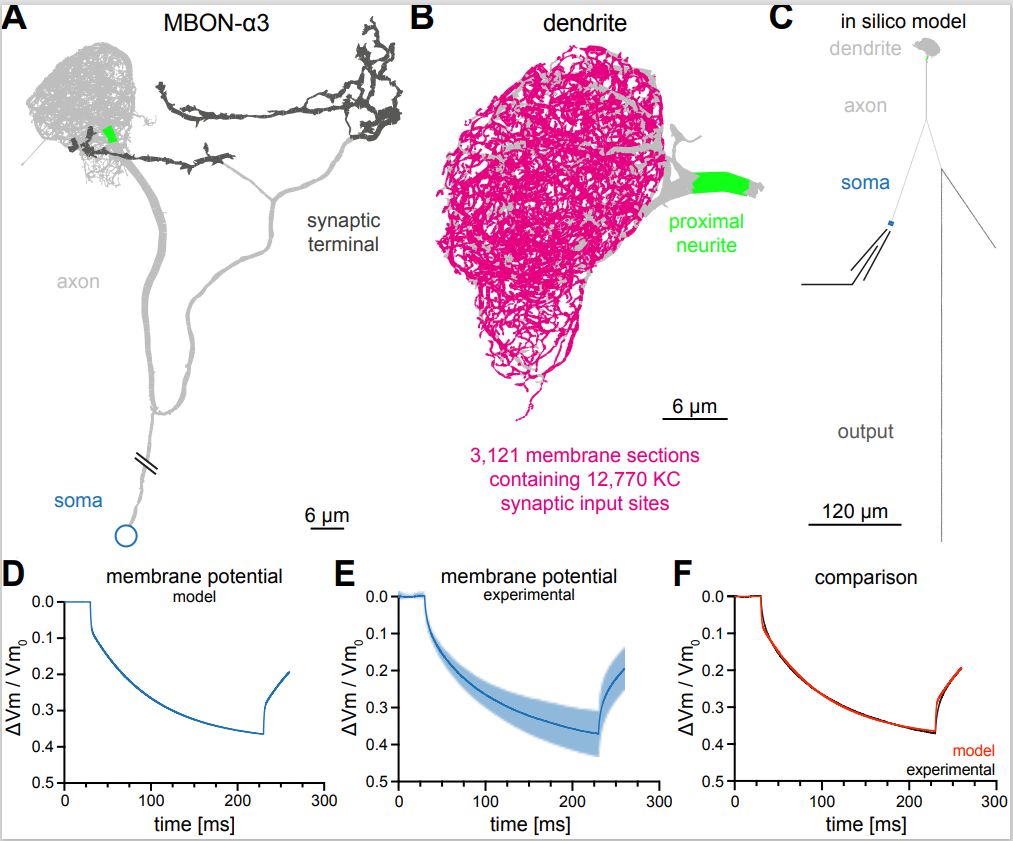
The Computational Neuroscience (CNS) Laboratory of the Mind/Brain Institute, headed by Prof. Ernst Niebur, just published in eLife a study using a new-to-the-MBI model organism: Drosophila melanogaster, the fruitfly. The paper investigated how sensory input leads to a decision on whether to flee from a dangerous or unpleasant environment. The investigators combined physiological results from their collaborators, Prof. Jan Pielage’s group from the University of Kaiserslautern, Germany, with a computational model of one of the key neurons in the circuit. The model was based on electron-microscopic data from colleagues at the HHMI Janelia Research Campus, in particular the groups of Drs. Yoshinori Aso and Glenn Turner that interact closely with the CNS lab. This anatomical detail allowed the model to incorporate each one of the thousands of synapses that provide the sensory input, each at their exact location on the neuron, and the branching structure of the neuron’s dendritic tree at the micrometer level. The results explain how this single neuron can efficiently store large numbers of memories using the observed flexible (stochastic) architecture of the circuit, allowing life-long learning of new memories by the animal.
These results open up the field for many new projects. The lab is actively searching for students to participate in these projects (contact: [email protected]).
Hafez et al (2023) The cellular architecture of memory modules in Drosophila supports stochastic input integration | eLife (elifesciences.org) 12:e77578.